Digital PCR (dPCR)

Digital PCR (dPCR) is a form of quantitative PCR based on end-point measurements. It has distinct advantages for specific types of assays:
- Absolute quantification
- Detection of rare target sequences
- Single cell analysis
- Detection of small fold changes (+/-10%)
- Quantification of proteins, PPIs and PTMs (QIAcuity system)
Lighthouse has a 6-color Stilla Naica Crystal Digital PCR System as well as a Qiagen QIAcuity system available for you to use. The Bio-Rad QX100 system has been retired.
When should I use Digital PCR?
Digital PCR excels in the measurement of low copy numbers, and has a linear range of 5 logs (from 1 to approximately 100,000 copies per reaction). The excellent linearity of the process is what allows the detection of very small fold-changes.
Because of its ability to absolutely quantify the number of molecules present within a sample, the use of a reference gene is not necessarily required for digital PCR. In addition, dPCR allows rare molecules to be detected within complex mixtures much more efficiently than standard QPCR. This is particularly useful, for example, in applications such as detecting circulating tumor DNA in cancer patients, where there are overall very few molecules of DNA to detect, within a large background of wild type DNA molecules.
Furthermore, dPCR is much more resistant to inhibitors than standard QPCR. Many reactions that do not work with QPCR can be amplified by dPCR. Similarly, setting up multiplexing reactions is much easier with dPCR than QPCR, again because the reactions are partitioned apart from each other. The Lighthouse Stilla system can analyze samples with up to 6 colors, making it suitable for multiplexing.
Digital PCR: How does it work?
Digital PCR works by "partitioning" or separating the QPCR reaction into many small reactions (slightly less than 1 nL volume for the Stilla). The Lighthouse system creates many small droplets within a water-in-oil emulsion, which are located within different regions or "wells" on a special optical slide.
|
|
Droplets are generated as a water-in-oil emulsion, in which the sample is partitioned into 20-30,000 aqueous droplets within an oil carrier. |
| The target and background DNA are randomly distributed throughout the droplets when they are formed. The sample is heated and cooled as for an ordinary PCR reaction. | |
 |
After the amplification step, each droplet provides a fluorescent positive or negative signal, indicating whether or not the target was present within the droplet after partitioning. |
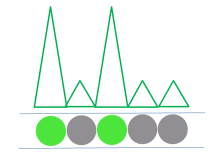 |
Droplets are examined individually, providing an independent digital measurement for each. Depending on their signal, the droplets are scored as either positive or negative for the marker. |
Based on the number of positive droplets and the number of partitions/droplets overall, the number of target molecules which were originally present in the reaction can be calculated with help of the Poisson distribution.
Stilla Naica System - Crystal Digital PCR
Lighthouse has a 6-color Stilla Naica Crystal Digital PCR System available for use.
On the Stilla system, the sample first flows through a network of microchannels and is partitioned into a large 2D array of up to 30,000 individual droplets, also called a droplet crystal. PCR is then performed on-chip and the droplets are scanned to reveal those which contain amplified targets. The last step consists of counting the number of these positive droplets to precisely extract the abolute quantity of nucleic acids.
Workflow:

1. Sample pipetting: Only one pipetting step is required to load the reaction mixes into the wells of the Sapphire (or Opal) Chips.
The wells are then sealed and the chip is placed into the Naica Geode.

2. Droplet Crystal Generation and Amplification: Once the chips are placed into the Naica Geode, launch the combined partitioning and thermocycling program. Crystals of up to 30,000 droplets are created from each sample, and PCR amplification is performed immediately after crystal generation.

3. Crystal reading: Transfer the chips to the Naica Prism6 instrument. Crystals are read using 3 fluorescence channels and the Crystal Reader program.
4. Data analysis: The Crystal Miner software automatically measures the concentrations of targeted nucleic acids. Analyze your data and explore your crystals using the program.
Primer/Probe Design for Digital PCR
The design of hydrolysis probes and primers is critical to the success of the experiment. The Core Facility has a copy of the Beacon Designer program to help with primer and probe design and seeing that these parameters are met.
Digital PCR: Compatible Mixes
|
System and Detection Method |
Example Fluorophores | Compatible Mixes |
| Stilla Naica (Probes) |
FAM Yakima Yellow (eq. HEX / VIC) Atto 550 / TAMRA ROX CY5 Atto 700 |
R10054 (small size, 5x mix, Buff.A) R10104 (small size, 10x mix, Buff. A) R10105 (large size, 10x mix, Buff. A) |
|
Stilla Naica (Evagreen) (similar to SYBR green) |
Evagreen |
R10056 (large, 5x Buff. A) R10106 (large, 10x Buff. A) |
|
Chips |
Stilla Sapphire Chips 4 samples / chip (12 chips/pk., 48 wells tot.) 25 µL/well, tot. rxn. vol. Cat. C14012 |
Stilla Ruby Chips 16 samples / chip (12 chips/pk., 192 wells tot.)
5 µL/well, total rxn. vol. Cat. C16011 |
The Naica mixes come in large and small size packages. The large package is for 300 Sapphire Chip and 1050 Opal chip reactions. The small package is for 150 Sapphire chip and 525 Opal chip reactions.
Packages with 10x Buffer A are recommended for samples where the maximum volume of DNA needs to be added (e.g. rare event detection, etc.).
Qiagen QIAcuity Digital PCR System
Lighthouse also has a Qiagen QIAcuity available for use. The QIAcuity is a nanoplate based system which uses a grid to form partitions within the wells, rather than droplets in an emulsion. The QIAcuity is a five-color system able to look at the same assortment of fluorophores as the Stilla Naica, with the exception of Atto 700.
The Qiagen system works with 24-well or 96-well nanoplates and is compatible with standard QPCR mixes.
| Nanoplate type | Partitions per Well | Total Rxn. Vol. | Catalog Number |
| 96-well | 8.5k | 12 µL | 250021 |
| 24-well | 8.5k | 12 µL | 250011 |
| 24-well | 26k | 40 µL | 250001 |
When used with Actome's Protein Interaction Coupling (PICO) Kits, the QIAcuity system also enables the analysis of proteins, protein interactions, and post-translational modifications from biological systems, low background and low sample input volume. For information about this application, please contact Actome (info@actome.de).
Some Useful Documents about dPCR:
|
User manual for the Naica Crystal Miner software |
|
|
User manual for the Naica Crystal Reader |
|
|
How to calculate Limit of Blank (LOB) and Limit of Detection (LOD) |
|
|
Bio-Rad technical note on detecting rare mutant alleles with ddPCR |
|
|
Bio-Rad application guide on using ddPCR for Rare Event Detection |
|
| Bio-Rad technical note on looking at copy number variation by ddPCR |
QPCR/dPCR Links of Interest
dPCR Calendar
Home


